Impact of the hockey stick spreading on microorganism enumeration
Whether you call it a hockey stick, a spreader, a spatula, the L-shaped thing for spreading or Drigalski’s rake (the debate is still open on LinkedIn), the use of this instrument must be mastered to perfection!
Use it too intensively and you run the risk of killing microorganisms; use it too “laxly” and enumeration will be impossible!
In short, spreading is not a trivial action.

What is the Drigalski spreader?

The Drigalski spatula was invented by… Wilhelm von Drigalski (a German bacteriologist). Originally, spatulas were made of glass or metal. They consisted of a handle and a triangle-shaped part (in contact with the agar).
Today’s spreader are more L-shaped and made of plastic. Some microbiology laboratories make them themselves by heating a Pasteur pipette… and yes, microbiologists can also be formidable glass sculptors.
In the end, whatever the shape or name, the use remains the same.
The spreader is a tool used by microbiologists not for gardening (though) but for spreading a solution of microorganisms on the surface of agar. During spreading, the liquid penetrates the agar, ensuring even distribution of the microorganisms. This manipulation facilitates the enumeration of microorganisms.
Because yes, the main purpose of the rake is to count the microorganisms present in the starting sample.
Best QC strains
Food & Water : The best QC Strain suppliers
We polled the SuperMicrobiologists.
Here are their favorite QC Strains
Beware of spreading time
A study by Hedderich et al* has shown that the duration of spreading can affect the viability of certain strains, particularly Gram-negative bacteria.
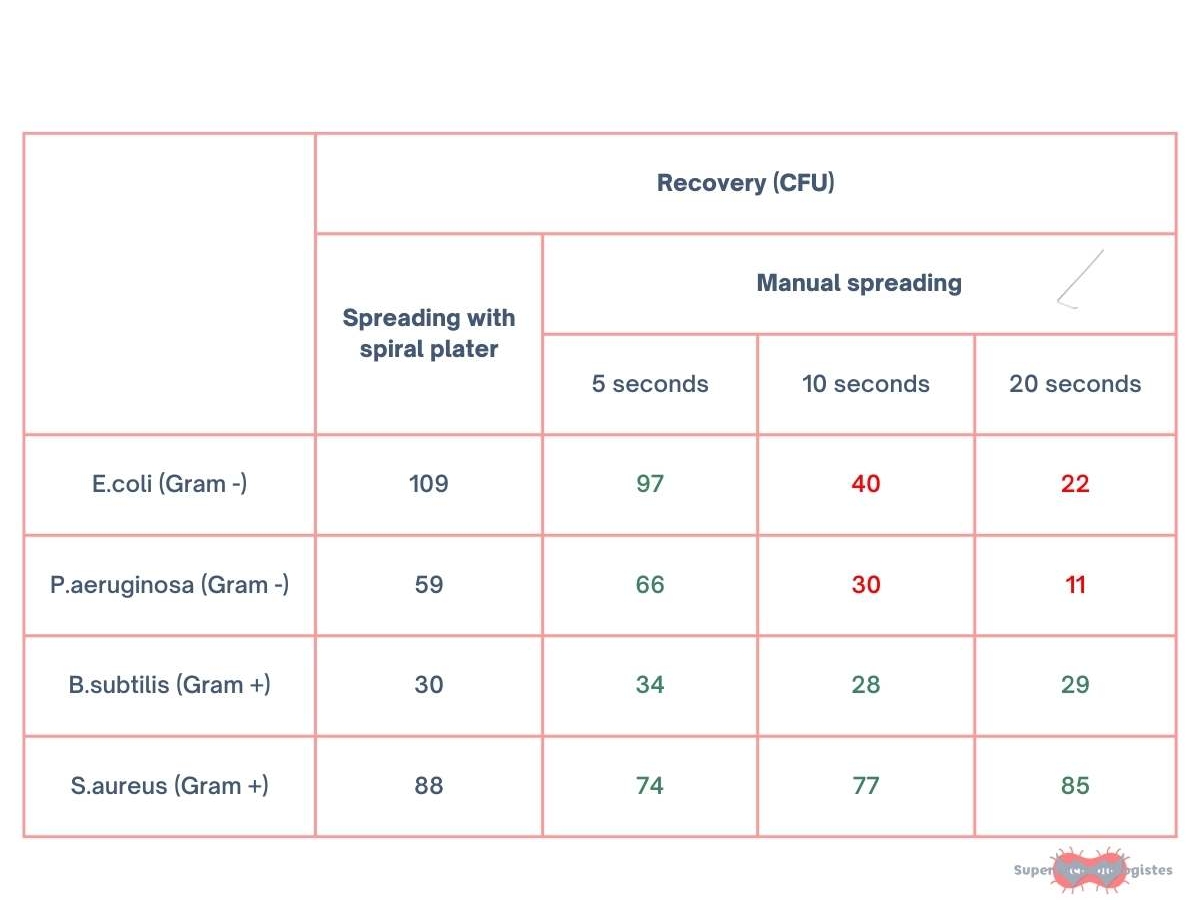
We found that 50% of Gram-negative bacteria (Escherichia coli and Pseudomonas aeruginosa) were lost after 10 seconds of spreading, and 80% after 20 seconds. Quite a shock, isn’t it?
The researchers then observed the state of the microorganisms with an electron microscope
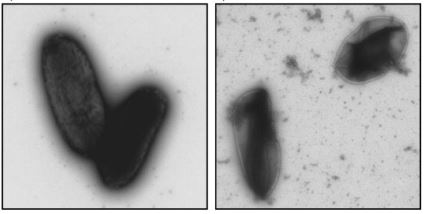
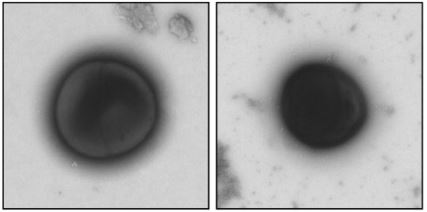
Electron microscope observations clearly show that raking causes severe damage to the membranes of Gram-negative bacteria. On the other hand, raking does not have the same effect on the membranes of Gram-positive bacteria.
Best QC strains
Pharma : The best QC Strain suppliers
We polled the SuperMicrobiologists.
Here are their favorite QC Strains
Conclusion
“Whether it’s for fertility testing or routine enumeration analysis, you’ve got to go easy on the hockey stick spreader.”
In addition to challenging culture media, fertility tests can also reveal a handling problem in the microbiology laboratory… and in our particular case, a spreading problem.
If during your fertility tests you regularly have recovery problems for your Gram-negative bacteria, it may be due to the length of the spread. After investigation, if spreading is indeed to blame, you’ll need to review your internal procedures to standardize this step, limiting it to 5 seconds or 5 “rakes”, for example.
But the most important thing is to discuss the matter with your microbiology colleagues, to find out about their practices and possibly alert them to this little-known problem.
- Hedderich, R., Müller, R., Greulich, Y., Bannert, N., Holland, G., Kaiser, P., Reissbrodt, R. Mechanical damage to Gram-negative bacteria by surface plating with the Drigalski-spatula technique.
(2011) International Journal of Food Microbiology, 146 (1), pp. 105-107.
After reading this article, if you have any objections or additional questions, please add to the debate by leaving a comment below.














Leave a Reply
Want to join the discussion?Feel free to contribute!