Why is STEC detection in food sample so complicated?
Whether it’s for the ISO/TS 13136 standard or for alternative methods, detecting strains of Escherichia coli that produce Shiga toxins (STEC) is quite a challenge.
Here’s why.
Content :
- Definition of a STEC
- Risk of false negatives
- Risk of false positives
- The presumptive positives
- Conclusion
The 6 best PCR for STEC
The 6 best method for STEC detection
We surveyed the SuperMicrobiologists,
here are the top-rated methods
What is a STEC?
We wrote a complete (and simple) article on the definition of STEC.
But in summary a STEC is an Escherichia coli with the stx gene, which is the gene that codes for the shiga toxins.
but if it came off that way, it would be (too) simple…
Indeed, not all the E.coli with an stx gene are pathogens. This is why in some countries, (usually where the prevalence of E.coli with stx genes is high), they go further in the definition of pathogenic STEC.
For example in France a HP STEC (Highly Pathogenic STEC) is an E.coli strains which has stx AND eae genes AND is part of the Big 5 (5 most dangerous E.coli serotypes for human beings).
So the first question to ask yourself is “what is considered as a pathogenic STEC for the countries where your products will be delivered”.
STECs are easily destroyed by heat. The main risk of contamination therefore concerns raw meat, milk, vegetables or undercooked products.
Risk of False Negatives in Detecting HP STEC

The risk of obtaining false-negative results is arguably the worst scenario for an industrial producer. Indeed, releasing a batch of contaminated products can impact consumer health and the company’s image.
This risk of false-negative results is significant when detecting HP STEC.
Why?
Simply because the matrices to be analyzed are complex (cheese, meats, plants). These matrices contain natural inhibitors (fat, physicochemical characteristics like pH) or interferents (ancillary flora) that could disrupt the detection of HP STEC by PCR.
Some alternative methods can mitigate the matrix effect (specific broth, IMS, etc.).
However, it’s essential to verify that the method has been validated with your matrices (as almost all matrices have their peculiarities, a small “in-house” validation is strongly recommended).
Risk of False Positives in Detecting HP STEC
A false positive means that the PCR detects a STEC marker when none are present in the sample. Given the sensitivity of PCR methods used today, this is unlikely. However, PCR could detect DNA from dead cells. Therefore, particular attention must be paid to this parameter when choosing a technology.
The validation file provided by the PCR supplier (or better an independent lab) could help you to better understand the sensitivity of the method.
The Confirmation of “Presumptive Positives”
(only if you are looking for stx+ AND/OR eae+ AND/OR specific serogroups)
This is arguably THE point that crystallizes all attention today. Here’s why with this example:
| Sample 1 | Sample 2 |


| PCR screening result: POSITIVE | PCR screening result: POSITIVE |
| Presence of HP STEC: yes | Presence of HP STEC: no |
| In this sample, there’s 1 bacteria possessing all markers (stx, eae, and the serotype O26:H11). After extraction, the genes are in the same suspension, it’s a presumptive positive sample. | In this sample, there are 3 bacterias, each carrying a different marker (stx, eae, and serotype O26: H11). After extraction, the genes are in the same suspension, it’s a presumptive positive sample |
In both cases, PCR screening will return a positive result*. However, only sample 1 is contaminated with an HP STEC.
That’s why, following the PCR screening step, confirmation through bacterial isolation on agar plates is mandatory. This verifies that the three markers come from the same bacteria.
The issue is that the presence of presumptive positives might not be rare in some matrices.
*When using the protocol described in the ISO/TS 13136 standard. Some alternative methods can reduce the percentage of presumptive positive samples.
Confirmation Techniques
Every PCR supplier proposes a specific protocol for confirmation. Although different, the general principle of these confirmation protocols is the same:
Isolate colonies to verify if the genes come from the same bacteria.
For confirmation, the enrichment broth is isolated on a specific culture media (chromogenic media or “selective” media such as TBX).
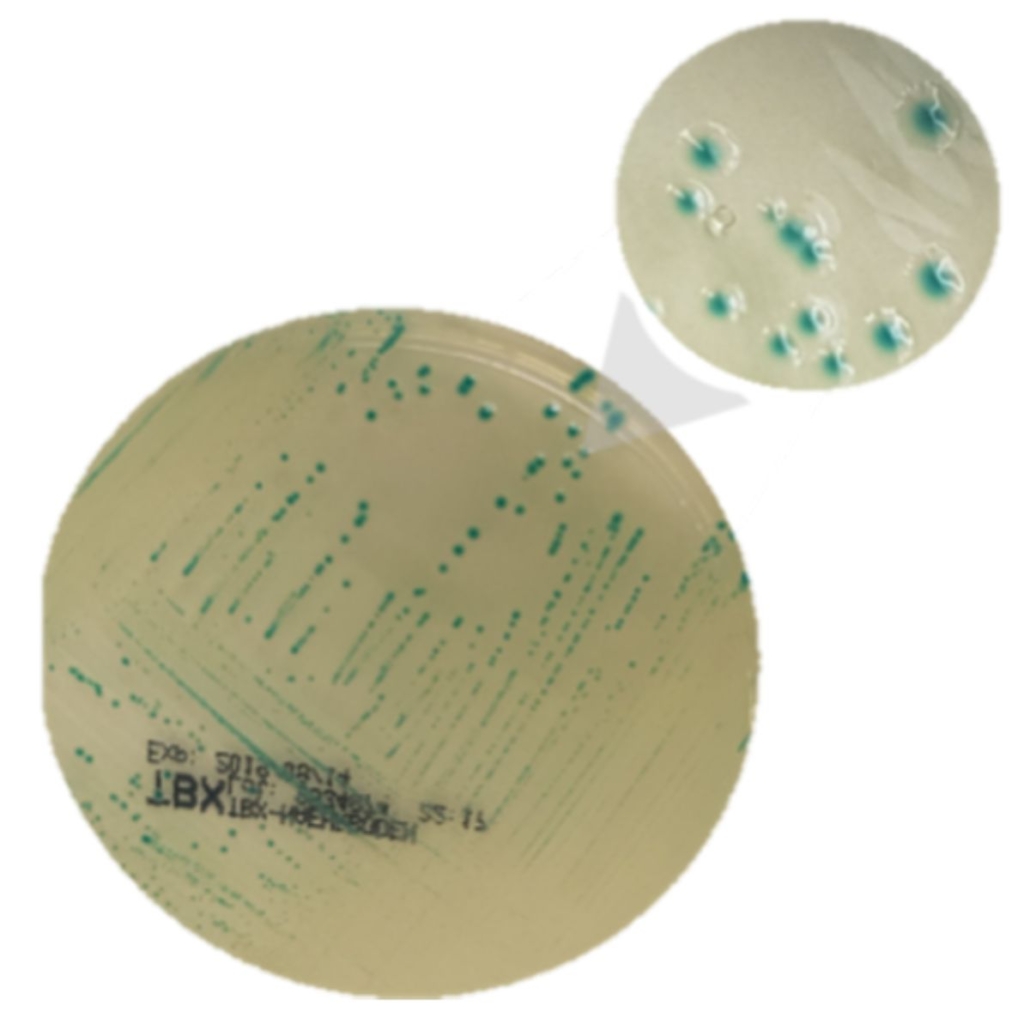
Once the colonies are visible, typical colonies are sampled and a PCR is conducted. If the genes (stx and eae) are present, it’s a positive STEC; otherwise, other colonies are sampled.
The ISO 13136 standard requires checking up to 50 colonies… that’s a lot of work, isn’t it?!
As we’ve just seen, this confirmation step is long, arduous, costly, and requires a certain level of expertise… It must not be forgotten that during this time, the sample is considered potentially positive, so it cannot be shipped (or if it already has been, it may be subject to recall). So there is a lot of pressure on the lab !
Therefore, we understand why the presence of presumptive positives during PCR screening can be a real problem for industrial producers.
Conclusion
The challenge for alternative methods will be to limit the number of presumptive positives while avoiding false negatives. We precisely address this topic in another dedicated article “How to Choose Your STEC Detection Method?“.















Trackbacks & Pingbacks
[…] explaining what STEC is, why they are complicated to detect, and how to choose the most suitable method for your lab, here is the final step: A selection of 6 […]
Leave a Reply
Want to join the discussion?Feel free to contribute!